R How to Modify Facet Plot Labels of ggplot2 Graph (Example Code)
In this R tutorial you’ll learn how to modify the facet labels of a ggplot2 facet plot.
Preparing the Example
We’ll use this example data:
data(iris) # Loading iris data head(iris) # Sepal.Length Sepal.Width Petal.Length Petal.Width Species # 1 5.1 3.5 1.4 0.2 setosa # 2 4.9 3.0 1.4 0.2 setosa # 3 4.7 3.2 1.3 0.2 setosa # 4 4.6 3.1 1.5 0.2 setosa # 5 5.0 3.6 1.4 0.2 setosa # 6 5.4 3.9 1.7 0.4 setosa |
data(iris) # Loading iris data head(iris) # Sepal.Length Sepal.Width Petal.Length Petal.Width Species # 1 5.1 3.5 1.4 0.2 setosa # 2 4.9 3.0 1.4 0.2 setosa # 3 4.7 3.2 1.3 0.2 setosa # 4 4.6 3.1 1.5 0.2 setosa # 5 5.0 3.6 1.4 0.2 setosa # 6 5.4 3.9 1.7 0.4 setosa
If we want to draw our data with the ggplot2 package, we have to install and load ggplot2:
install.packages("ggplot2") # Install ggplot2 package library("ggplot2") # Load ggplot2 package |
install.packages("ggplot2") # Install ggplot2 package library("ggplot2") # Load ggplot2 package
ggplot(iris, aes(x = Sepal.Length, # Plotting ggplot2 facet graph y = Petal.Length)) + geom_point() + facet_grid(Species ~ .) |
ggplot(iris, aes(x = Sepal.Length, # Plotting ggplot2 facet graph y = Petal.Length)) + geom_point() + facet_grid(Species ~ .)
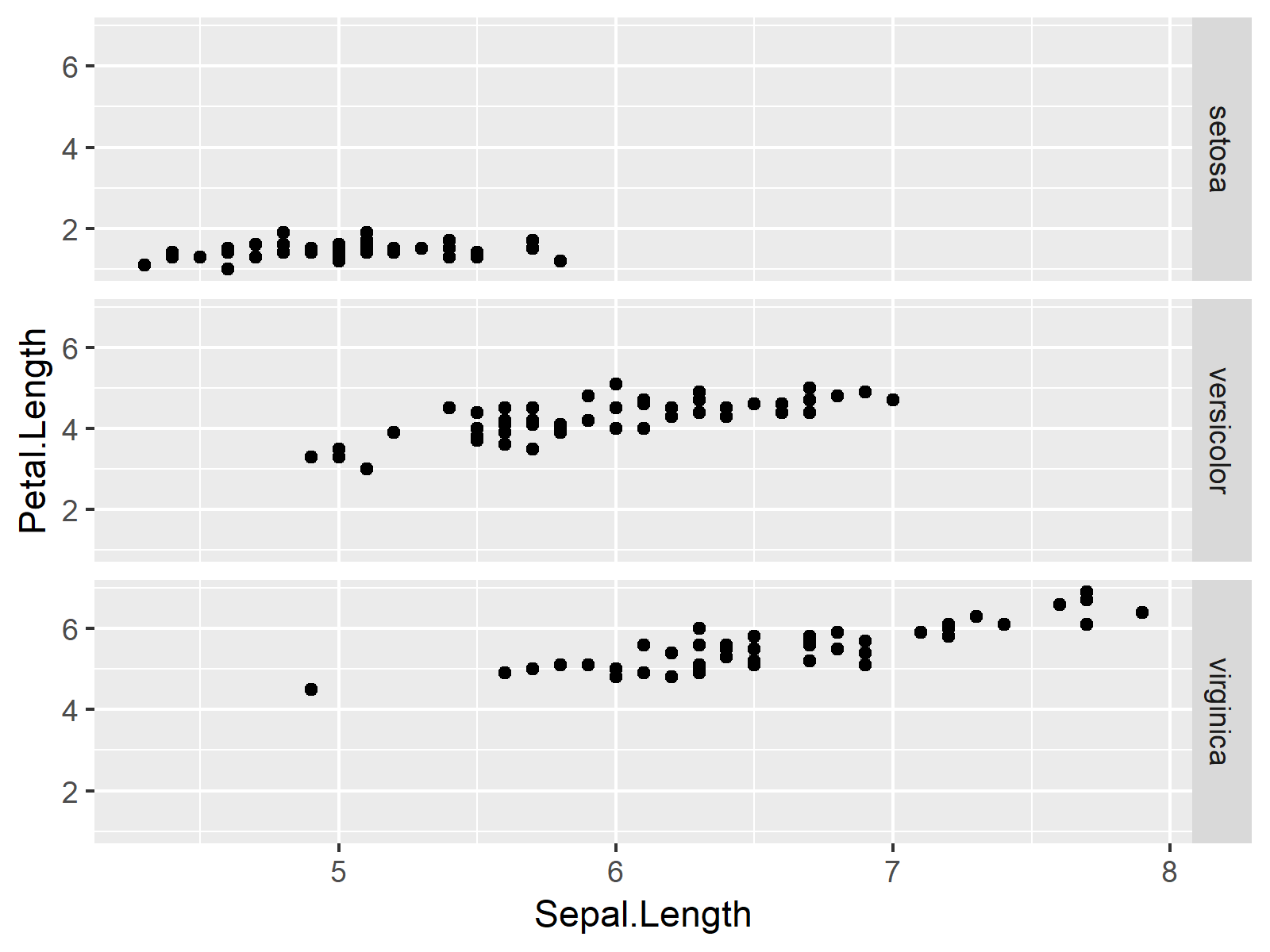
Example: Print Different ggplot2 Facet Plot Labels by Changing Factor Levels
iris_new <- iris # Duplicating data frame levels(iris_new$Species) <- c("Species No. 1", # Adjusting Species factor levels "Species No. 2", "Species No. 3") |
iris_new <- iris # Duplicating data frame levels(iris_new$Species) <- c("Species No. 1", # Adjusting Species factor levels "Species No. 2", "Species No. 3")
ggplot(iris_new, aes(x = Sepal.Length, # Plotting ggplot2 facet graph y = Petal.Length)) + geom_point() + facet_grid(Species ~ .) |
ggplot(iris_new, aes(x = Sepal.Length, # Plotting ggplot2 facet graph y = Petal.Length)) + geom_point() + facet_grid(Species ~ .)
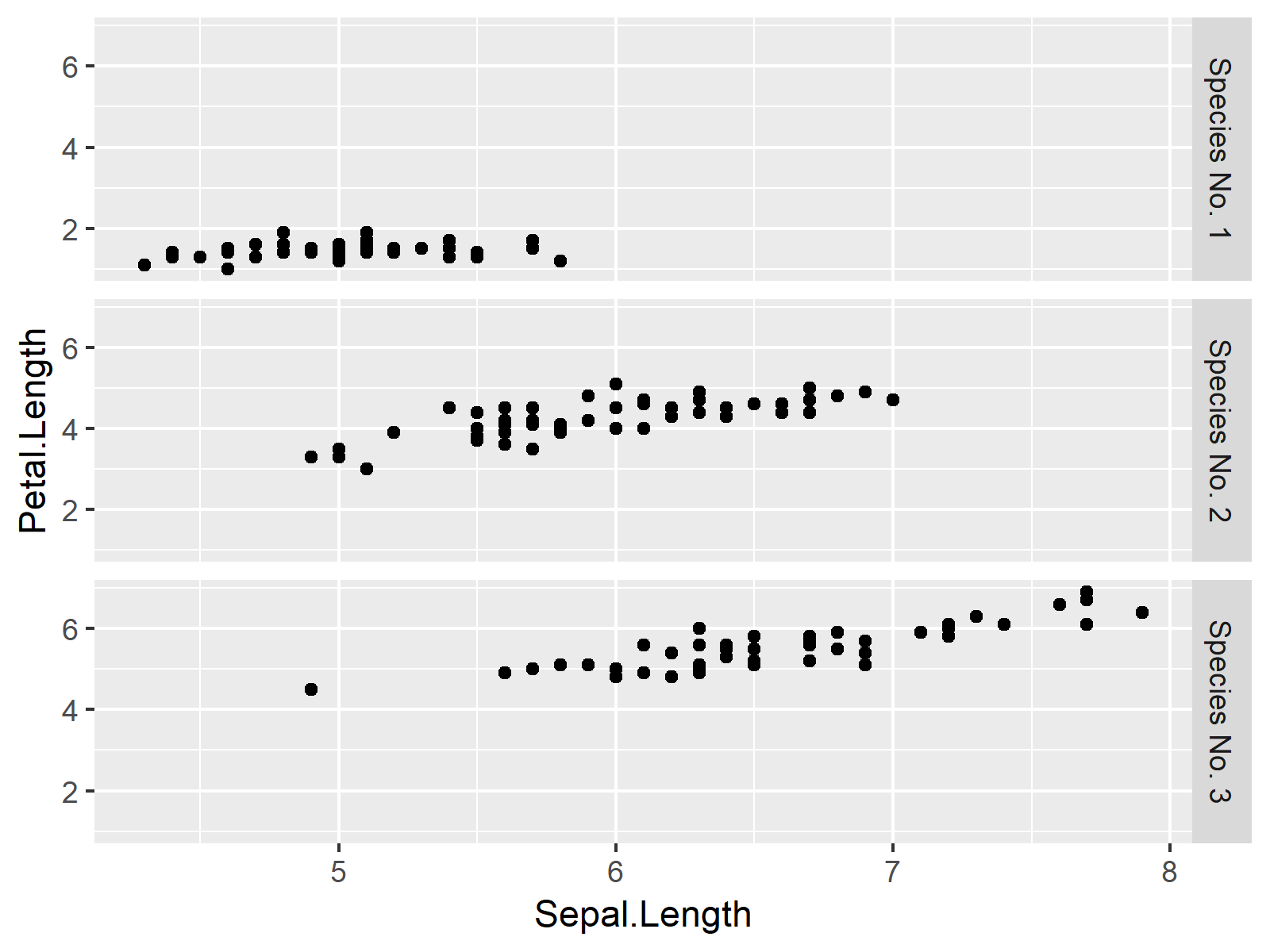
Related Tutorials
In the following, you may find some further resources on topics such as graphics in r, lines, colors, and plot legends.
