Change Alpha of Points in Plot – Base R & ggplot2 (2 Examples)
In this article you’ll learn how to decrease the alpha of points in an xy-plot using Base R and the ggplot2 package in the R programming language.
Preparing the Examples
data(iris) # Loading iris data set head(iris) # Sepal.Length Sepal.Width Petal.Length Petal.Width Species # 1 5.1 3.5 1.4 0.2 setosa # 2 4.9 3.0 1.4 0.2 setosa # 3 4.7 3.2 1.3 0.2 setosa # 4 4.6 3.1 1.5 0.2 setosa # 5 5.0 3.6 1.4 0.2 setosa # 6 5.4 3.9 1.7 0.4 setosa |
data(iris) # Loading iris data set head(iris) # Sepal.Length Sepal.Width Petal.Length Petal.Width Species # 1 5.1 3.5 1.4 0.2 setosa # 2 4.9 3.0 1.4 0.2 setosa # 3 4.7 3.2 1.3 0.2 setosa # 4 4.6 3.1 1.5 0.2 setosa # 5 5.0 3.6 1.4 0.2 setosa # 6 5.4 3.9 1.7 0.4 setosa
Example 1: Decrease Alpha in Base R Scatterplot
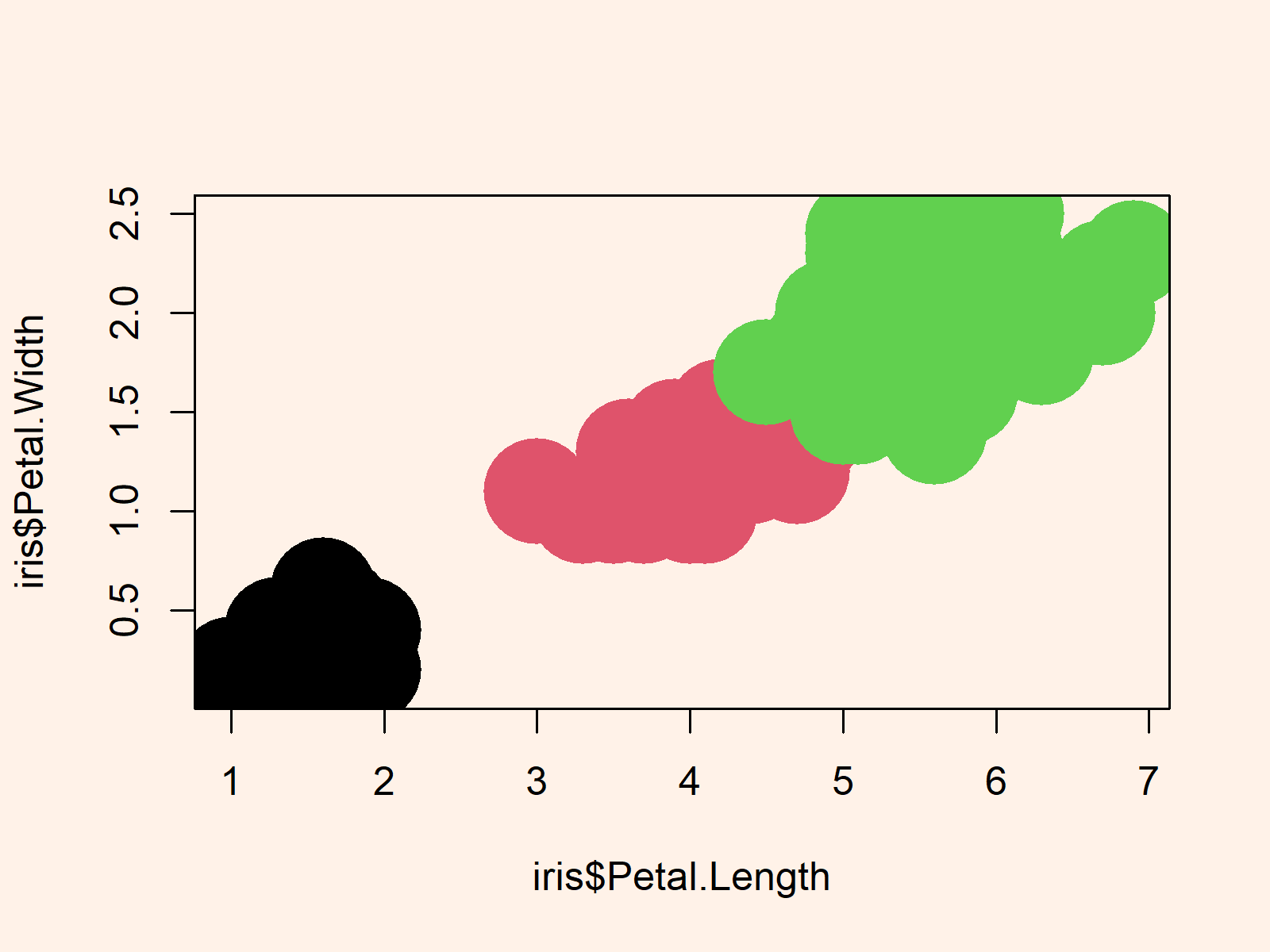
plot(iris$Petal.Length, # Graph without transparency iris$Petal.Width, cex = 6, pch = 16, col = iris$Species) |
plot(iris$Petal.Length, # Graph without transparency iris$Petal.Width, cex = 6, pch = 16, col = iris$Species)
install.packages("scales") # Install & load scales library("scales") |
install.packages("scales") # Install & load scales library("scales")
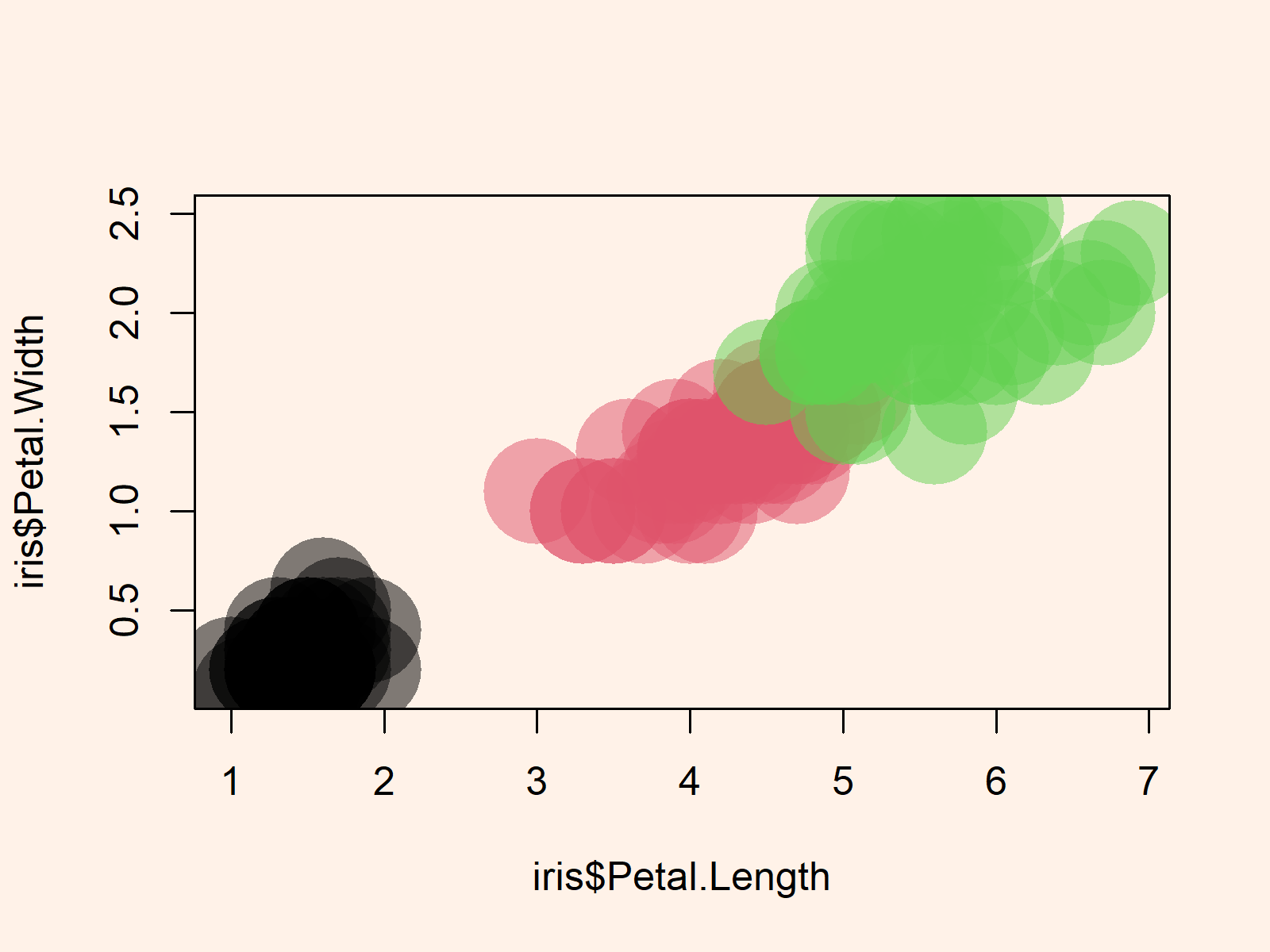
plot(iris$Petal.Length, # Transparent graph iris$Petal.Width, cex = 6, pch = 16, col = alpha(as.numeric(iris$Species), 0.5)) |
plot(iris$Petal.Length, # Transparent graph iris$Petal.Width, cex = 6, pch = 16, col = alpha(as.numeric(iris$Species), 0.5))
Example 2: Decrease Alpha in ggplot2 Scatterplot
install.packages("ggplot2") # Install & load ggplot2 library("ggplot2") |
install.packages("ggplot2") # Install & load ggplot2 library("ggplot2")
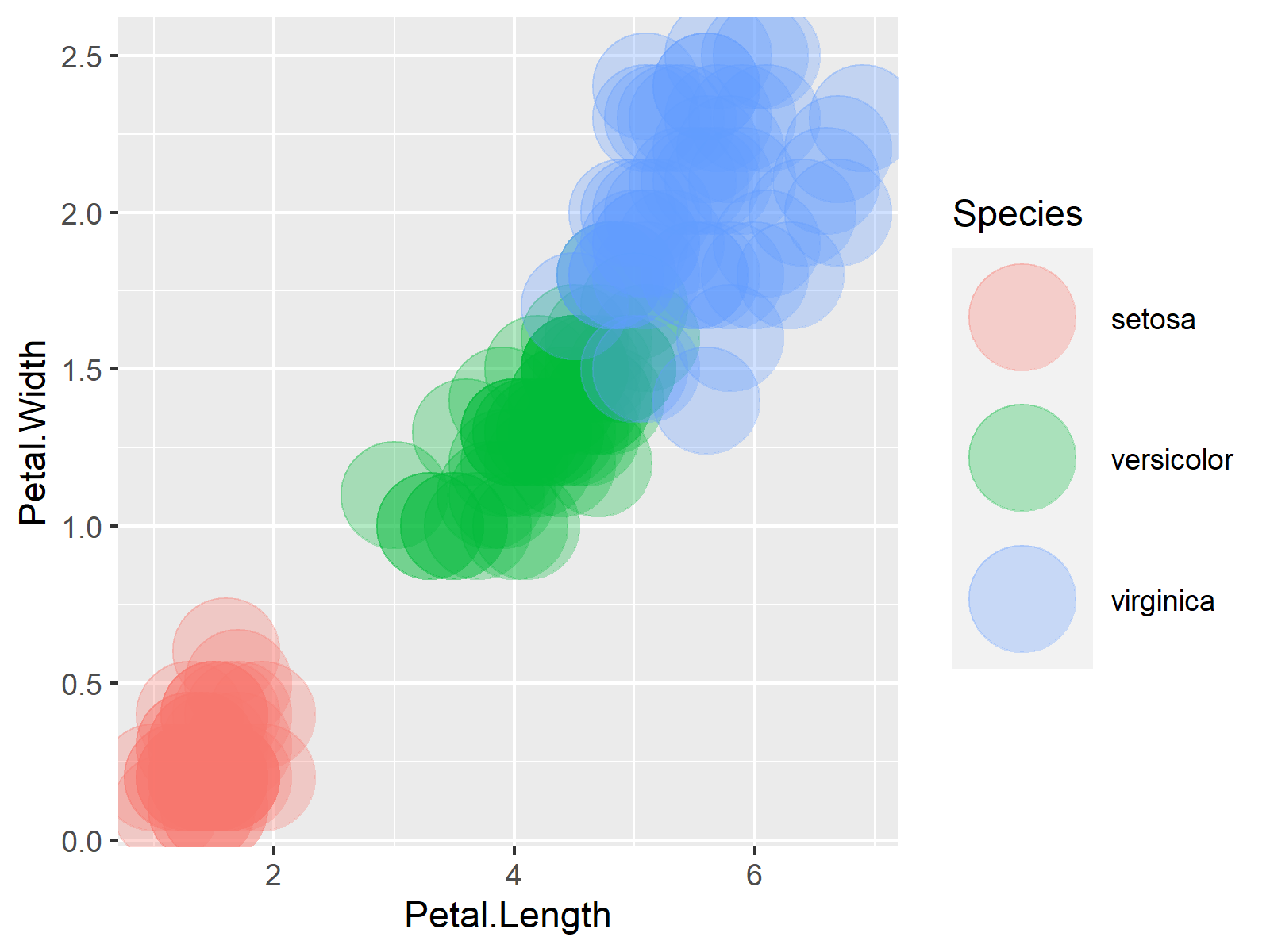
ggplot(iris, aes(Petal.Length, Petal.Width, col = Species)) + # Graph without transparency geom_point(size = 15) |
ggplot(iris, aes(Petal.Length, Petal.Width, col = Species)) + # Graph without transparency geom_point(size = 15)
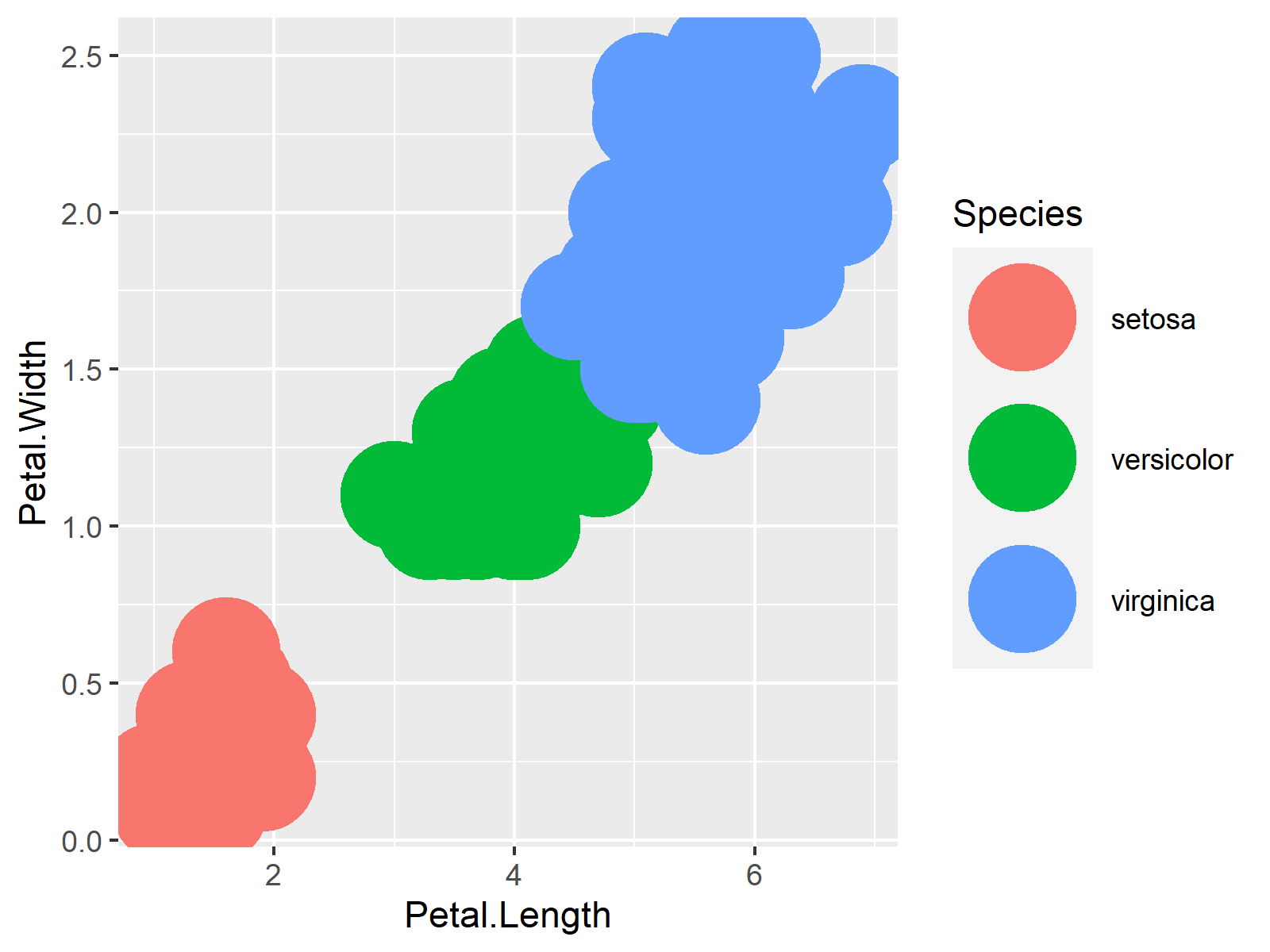
ggplot(iris, aes(Petal.Length, Petal.Width, col = Species)) + # Transparent graph geom_point(size = 15, alpha = 0.3) |
ggplot(iris, aes(Petal.Length, Petal.Width, col = Species)) + # Transparent graph geom_point(size = 15, alpha = 0.3)