How to Draw All Variables of a Data Frame in a ggplot2 Plot in R (Example Code)
In this article you’ll learn how to draw each column of a data matrix in a graphic in the R programming language.
Setting up the Example
my_df <- data.frame(time = 1:5, # Example data x1 = 5:1, x2 = 3:7, x3 = c(4, 1, 6, 5, 5)) my_df # Return data # time x1 x2 x3 # 1 1 5 3 4 # 2 2 4 4 1 # 3 3 3 5 6 # 4 4 2 6 5 # 5 5 1 7 5 |
my_df <- data.frame(time = 1:5, # Example data x1 = 5:1, x2 = 3:7, x3 = c(4, 1, 6, 5, 5)) my_df # Return data # time x1 x2 x3 # 1 1 5 3 4 # 2 2 4 4 1 # 3 3 3 5 6 # 4 4 2 6 5 # 5 5 1 7 5
my_df_reshaped <- data.frame(time = my_df$time, # Reshape data values = c(my_df$x1, my_df$x2, my_df$x3), col = c(rep("x1", nrow(my_df)), rep("x2", nrow(my_df)), rep("x3", nrow(my_df)))) my_df_reshaped # Return reshaped data # time values col # 1 1 5 x1 # 2 2 4 x1 # 3 3 3 x1 # 4 4 2 x1 # 5 5 1 x1 # 6 1 3 x2 # 7 2 4 x2 # 8 3 5 x2 # 9 4 6 x2 # 10 5 7 x2 # 11 1 4 x3 # 12 2 1 x3 # 13 3 6 x3 # 14 4 5 x3 # 15 5 5 x3 |
my_df_reshaped <- data.frame(time = my_df$time, # Reshape data values = c(my_df$x1, my_df$x2, my_df$x3), col = c(rep("x1", nrow(my_df)), rep("x2", nrow(my_df)), rep("x3", nrow(my_df)))) my_df_reshaped # Return reshaped data # time values col # 1 1 5 x1 # 2 2 4 x1 # 3 3 3 x1 # 4 4 2 x1 # 5 5 1 x1 # 6 1 3 x2 # 7 2 4 x2 # 8 3 5 x2 # 9 4 6 x2 # 10 5 7 x2 # 11 1 4 x3 # 12 2 1 x3 # 13 3 6 x3 # 14 4 5 x3 # 15 5 5 x3
install.packages("ggplot2") # Install & load ggplot2 package library("ggplot2") |
install.packages("ggplot2") # Install & load ggplot2 package library("ggplot2")
Example: Plot All Columns of Data Set with ggplot2 Package
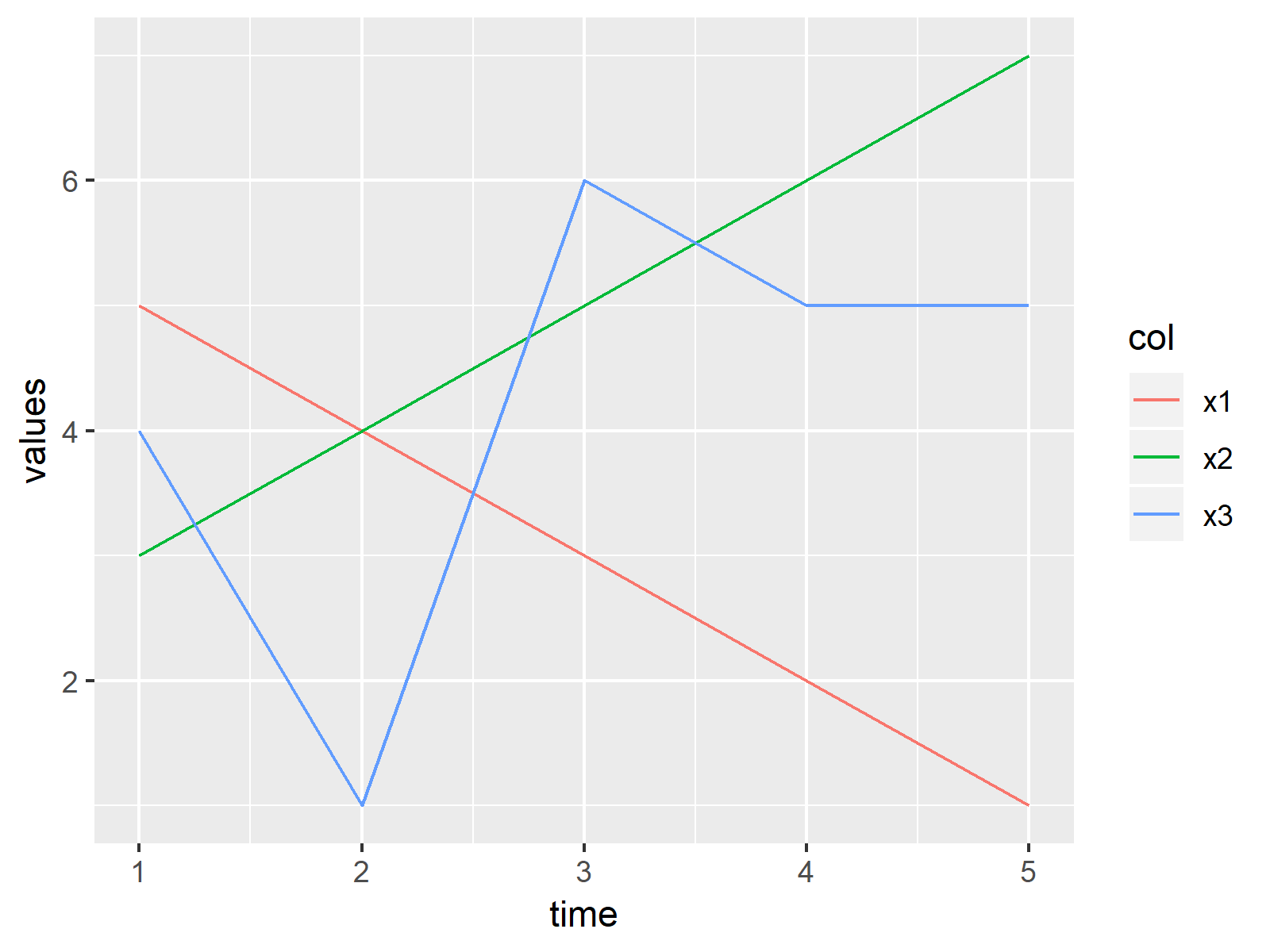
ggplot(my_df_reshaped, aes(x = time, y = values, col = col)) + # Drawing ggplot2 graph geom_line() |
ggplot(my_df_reshaped, aes(x = time, y = values, col = col)) + # Drawing ggplot2 graph geom_line()
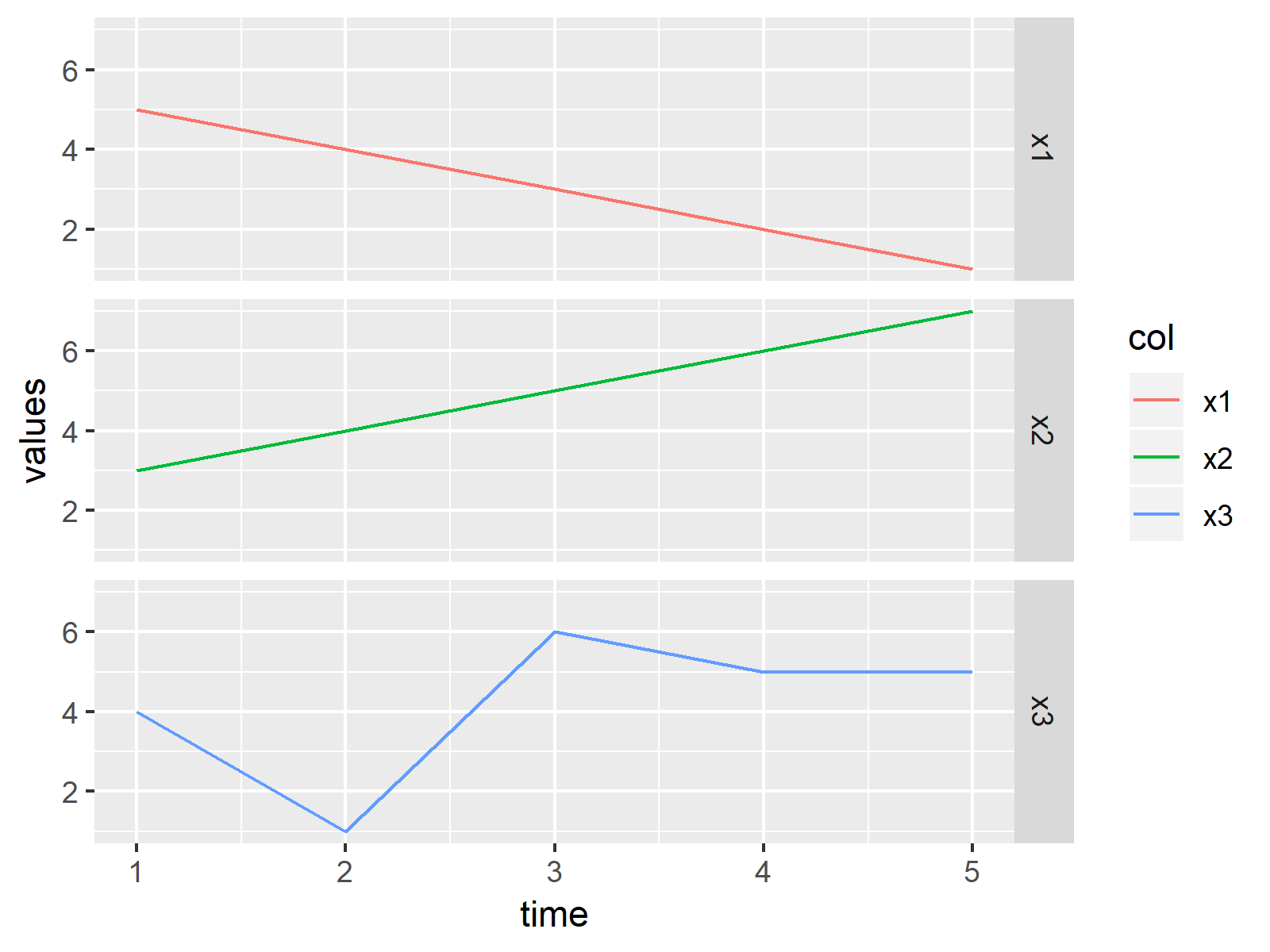
ggplot(my_df_reshaped, aes(x = time, y = values, col = col)) + # Drawing ggplot2 facet graph geom_line() + facet_grid(col ~ .) |
ggplot(my_df_reshaped, aes(x = time, y = values, col = col)) + # Drawing ggplot2 facet graph geom_line() + facet_grid(col ~ .)