Getting Started With data.table in R (6 Examples)
In this article, I’ll illustrate how to use the package data.table in R programming with several examples. For more in depth information on data.tables, we recommend you to take a look at the documentation on github and CRAN.
Setting up the Examples
We first install and load the package.
install.packages("data.table") # Install data.table package library("data.table") # Load data.table |
install.packages("data.table") # Install data.table package library("data.table") # Load data.table
We use the iris dataset to demonstrate the usage of the package. The structure of the original data is a data.frame which we convert into a data.table.
data(iris) # Load iris data set iris_DT_1 <- data.table::copy(iris) # Replicate the iris data set iris_DT_1 <- setDT(iris_DT_1) # Convert to data.table |
data(iris) # Load iris data set iris_DT_1 <- data.table::copy(iris) # Replicate the iris data set iris_DT_1 <- setDT(iris_DT_1) # Convert to data.table
head(iris_DT_1) # Printing the data head # Sepal.Length Sepal.Width Petal.Length Petal.Width Species # 1: 5.1 3.5 1.4 0.2 setosa # 2: 4.9 3.0 1.4 0.2 setosa # 3: 4.7 3.2 1.3 0.2 setosa # 4: 4.6 3.1 1.5 0.2 setosa # 5: 5.0 3.6 1.4 0.2 setosa # 6: 5.4 3.9 1.7 0.4 setosa |
head(iris_DT_1) # Printing the data head # Sepal.Length Sepal.Width Petal.Length Petal.Width Species # 1: 5.1 3.5 1.4 0.2 setosa # 2: 4.9 3.0 1.4 0.2 setosa # 3: 4.7 3.2 1.3 0.2 setosa # 4: 4.6 3.1 1.5 0.2 setosa # 5: 5.0 3.6 1.4 0.2 setosa # 6: 5.4 3.9 1.7 0.4 setosa
Above, you see the first data rows of the resulting data.table iris_DT_1.
Example 1: Choosing Specific Data Columns
There are different ways to address a column of a data.table. In the following, we display four different options.
In options 1 and 2, we use the name of the column Species to address it, either with the $ sign or via the indexing brackets [ , ]. Note that both ways, the values of the column are returned as a vector.
head(iris_DT_1$Species) # Option 1 # [1] setosa setosa setosa setosa setosa setosa # Levels: setosa versicolor virginica |
head(iris_DT_1$Species) # Option 1 # [1] setosa setosa setosa setosa setosa setosa # Levels: setosa versicolor virginica
In contrast to a data.frame, we do not need quotation marks around the column names within brackets [ , ].
head(iris_DT_1[, Species]) # Option 2 # [1] setosa setosa setosa setosa setosa setosa # Levels: setosa versicolor virginica |
head(iris_DT_1[, Species]) # Option 2 # [1] setosa setosa setosa setosa setosa setosa # Levels: setosa versicolor virginica
Alternatively, we can also address a column by its index or with its name stored as a character in another object, as shown in options 3 and 4. Both options return a data.table with a single column, not a vector.
head(iris_DT_1[, 5]) # Option 3 # Species # 1: setosa # 2: setosa # 3: setosa # 4: setosa # 5: setosa # 6: setosa |
head(iris_DT_1[, 5]) # Option 3 # Species # 1: setosa # 2: setosa # 3: setosa # 4: setosa # 5: setosa # 6: setosa
It might be a bit confusing when starting to work with data.table, but one gets used to it quickly: .SD, more info here. We can simply use it to index columns when their names are stored in other objects like name_of_selected_column.
name_of_selected_column <- "Species" head(iris_DT_1[, .SD, .SDcols = name_of_selected_column]) # Option 4 # Species # 1: setosa # 2: setosa # 3: setosa # 4: setosa # 5: setosa # 6: setosa |
name_of_selected_column <- "Species" head(iris_DT_1[, .SD, .SDcols = name_of_selected_column]) # Option 4 # Species # 1: setosa # 2: setosa # 3: setosa # 4: setosa # 5: setosa # 6: setosa
Note that we can use the same function, when name_of_selected_column is a vector containing several column names.
Example 2: Sub-Setting the iris data.table
In this example, we show how to filter all data rows of a data.table for which certain conditions hold. That is, we want to get all data rows of iris_DT_1 in which column Species takes level Setosa and column Sepal.Length takes values greater 5.
iris_DT_2 <- iris_DT_1[ Species == "setosa" & Sepal.Length > 5, ] # Data subset head(iris_DT_2) # Print head of the data # Sepal.Length Sepal.Width Petal.Length Petal.Width Species # 1: 5.1 3.5 1.4 0.2 setosa # 2: 5.4 3.9 1.7 0.4 setosa # 3: 5.4 3.7 1.5 0.2 setosa # 4: 5.8 4.0 1.2 0.2 setosa # 5: 5.7 4.4 1.5 0.4 setosa # 6: 5.4 3.9 1.3 0.4 setosa |
iris_DT_2 <- iris_DT_1[ Species == "setosa" & Sepal.Length > 5, ] # Data subset head(iris_DT_2) # Print head of the data # Sepal.Length Sepal.Width Petal.Length Petal.Width Species # 1: 5.1 3.5 1.4 0.2 setosa # 2: 5.4 3.9 1.7 0.4 setosa # 3: 5.4 3.7 1.5 0.2 setosa # 4: 5.8 4.0 1.2 0.2 setosa # 5: 5.7 4.4 1.5 0.4 setosa # 6: 5.4 3.9 1.3 0.4 setosa
Example 3: Getting the Number of Rows for Certain Data Subsets
We cannot only extract data subsets within the brackets [ , ], but also perform computations and define new columns within these. One particular useful option is to use .N to count the number of rows for which certain conditions hold. For that, within brackets [ , ] we condition the rows for which we want to count the number of occasions.
iris_DT_1[ Species == "virginica" & Petal.Length >= 5.7, .N] # [1] 19 |
iris_DT_1[ Species == "virginica" & Petal.Length >= 5.7, .N] # [1] 19
There are 19 data rows of iris_DT_1 in which column Species takes level virginica and column Petal.Length takes values greater or equal 5.7.
The number of counts can also be used to define additional data columns. In the following, we define an additional column named n_obs. It is defined as the number of counts (:= .N), where the counts are calculated for each species (, by = Species). That is, the entries of the same species are all assigned the same value of n_obs.
iris_DT_3 <- data.table::copy(iris_DT_1) # Replicate the data iris_DT_3[ , "n_obs" := .N, by = Species] # Define new column n_obs head(iris_DT_3) # Sepal.Length Sepal.Width Petal.Length Petal.Width Species n_obs # 1: 5.1 3.5 1.4 0.2 setosa 50 # 2: 4.9 3.0 1.4 0.2 setosa 50 # 3: 4.7 3.2 1.3 0.2 setosa 50 # 4: 4.6 3.1 1.5 0.2 setosa 50 # 5: 5.0 3.6 1.4 0.2 setosa 50 # 6: 5.4 3.9 1.7 0.4 setosa 50 |
iris_DT_3 <- data.table::copy(iris_DT_1) # Replicate the data iris_DT_3[ , "n_obs" := .N, by = Species] # Define new column n_obs head(iris_DT_3) # Sepal.Length Sepal.Width Petal.Length Petal.Width Species n_obs # 1: 5.1 3.5 1.4 0.2 setosa 50 # 2: 4.9 3.0 1.4 0.2 setosa 50 # 3: 4.7 3.2 1.3 0.2 setosa 50 # 4: 4.6 3.1 1.5 0.2 setosa 50 # 5: 5.0 3.6 1.4 0.2 setosa 50 # 6: 5.4 3.9 1.7 0.4 setosa 50
Example 4: Calculation of Statistics for Specific Subsets of the iris Data
Within the brackets [ , ], we can not only count instances, but basically perform any desired function, for a chosen set of data rows and columns. In the following example, we calculate the summary statistics of the quotient of columns Petal.Length and Petal.Width for those data rows for which Species is virginica.
iris_DT_1[ Species == "virginica", summary(Petal.Length / Petal.Width)] # Calculate summary statistics # Min. 1st Qu. Median Mean 3rd Qu. Max. # 2.125 2.511 2.667 2.781 3.056 4.000 |
iris_DT_1[ Species == "virginica", summary(Petal.Length / Petal.Width)] # Calculate summary statistics # Min. 1st Qu. Median Mean 3rd Qu. Max. # 2.125 2.511 2.667 2.781 3.056 4.000
Example 5: Calculation of Statistics by Groups
In this example, we calculate the mean value of column Petal.Length and the sum of column Petal.Width, for the different levels of column Species (by = Species). When we calculate several statistics, we have to use list() for these.
iris_DT_1[ , list("mean_Pet.L" = mean(Petal.Length), "sum_Petal.W" = sum(Petal.Width)), by = Species] # Species mean_Pet.L sum_Petal.W # 1: setosa 1.462 12.3 # 2: versicolor 4.260 66.3 # 3: virginica 5.552 101.3 |
iris_DT_1[ , list("mean_Pet.L" = mean(Petal.Length), "sum_Petal.W" = sum(Petal.Width)), by = Species] # Species mean_Pet.L sum_Petal.W # 1: setosa 1.462 12.3 # 2: versicolor 4.260 66.3 # 3: virginica 5.552 101.3
In the same manner, we can apply any other function to data subsets within a data.table.
Example 6: Creating Plots Within data.table
The applicability of functions within brackets [ , ] is not limited to statistics. For example, we can use it to create plots of data.table information as shown in the following.
iris_DT_1[ Sepal.Length >= 5 , plot(Petal.Length, Petal.Width, pch = 20, col = "blue") ] |
iris_DT_1[ Sepal.Length >= 5 , plot(Petal.Length, Petal.Width, pch = 20, col = "blue") ]
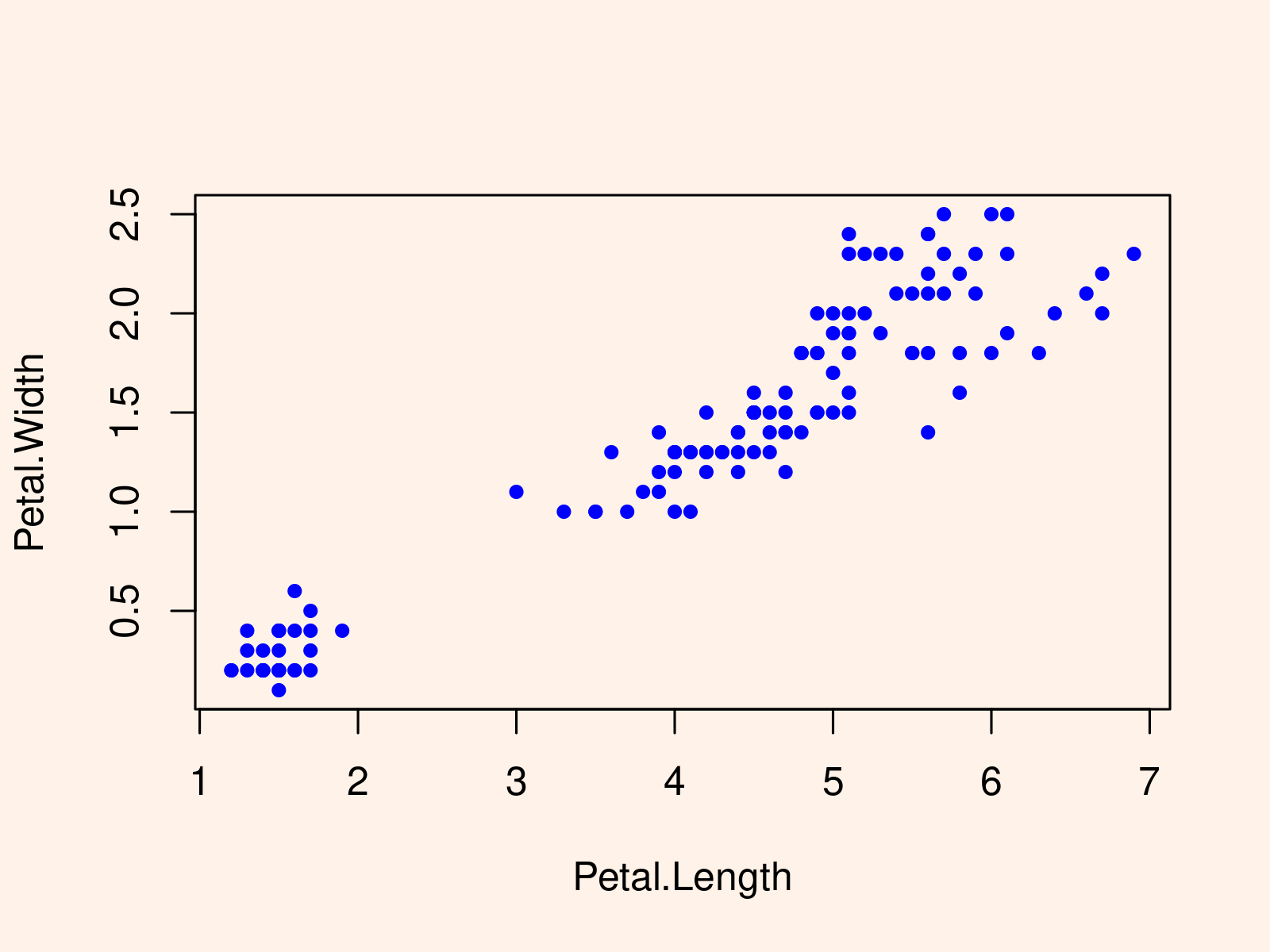
The scatterplot shows the values of Petal.Length and Petal.Width for all those data rows in which Sepal.Length is greater or equal to 5.
Note: This article was created in collaboration with Anna-Lena Wölwer. Anna-Lena is a researcher and programmer who creates tutorials on statistical methodology as well as on the R programming language. You may find more info about Anna-Lena and her other articles on her profile page.

